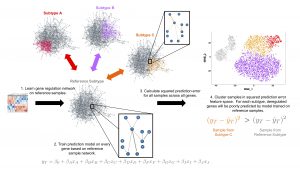
We reached an important milestone today. Kristina got a major part of her thesis work accepted for publication in the journal Bioinformatics. The paper describes a new method, single sample Network Perturbation Assessment (ssNPA), which learns a gene expression network (in the form of a directed graph) from a set of reference samples. Then for every query sample, it evaluates which network neighborhoods are perturbed and by how much. The resulting perturbation features can be used to subsequently cluster the non-reference samples.
We reached an important milestone today. Kristina got a major part of her thesis work accepted for publication in the journal Bioinformatics. The paper describes a new method, single sample Network Perturbation Assessment (ssNPA), which learns a gene expression network (in the form of a directed graph) from a set of reference samples. Then for every query sample, it evaluates which network neighborhoods are perturbed and by how much. The resulting perturbation features can be used to subsequently cluster the non-reference samples.
The title of the paper is “Causal network perturbations for instance-specific analysis of single cell and disease samples” and it will soon be published on the journal’s web site.
A pre-print of this paper can be found on bioRxiv (here). [Uploaded on May 16, 2019]
The code for ssNPA and the datasets used in the paper can be found here.
Congratulations Kristina!
[UPDATE 24-Dec-2019] The paper is now published at the Bioinformatics web site. (here)